| News: |
2022-06-07 -- Preprint on BC Hub deposited in bioRχiv
|
|
| Latest publication: Ambrosini, G., Agnoletto, A., Brisken, C., Bucher, P. The Breast Cancer Epigenomics Track Hub. bioRxiv; 2022. doi: 10.1101/2022.05.01.490187. | ||
Quick Information for New Users
- Have a look at the Beginner's Tutorial to find out what you can do with this resource.
- Visit our Session Gallery to learn about epigenomics data by exporing interesting cases.
- Check out the Table below to see what's currently available.
- Read our Citation Instructions.
Breast Cancer Epigenomics Track Hub
The Breast Cancer (BC) hub and viewer is a tool meant to provide a new way to explore cancer-related epigenomic data using the UCSC genome browser as a visualization platform, aiming at making omics data FAIR: Findable, Accessible, Interoperable and Reusable.
The hub is a collection of public high-resolution tracks from different labs, whose public data is downloaded from primary sources, such as GEO.
The access button and the links posted below automatically connect the user to other public track hubs (e. g. the Cancer Genome Atlas hub) in addition to relevant tracks from the UCSC genome browser database.
Such simultaneous view of data from different large-scale cancer studies helps identify patterns of epigenomic gene regulation.
The prospective user community are biologists interested in a specific gene regulatory region. The BC hub is designed to make it easier to users to find and assemble relevant data for visual inspection of such regions.
Access to the Hub
To access the BC hub please click on the button here :
Here below, we provide links to a series of browser screenshots (sessions) related to interesting use cases.
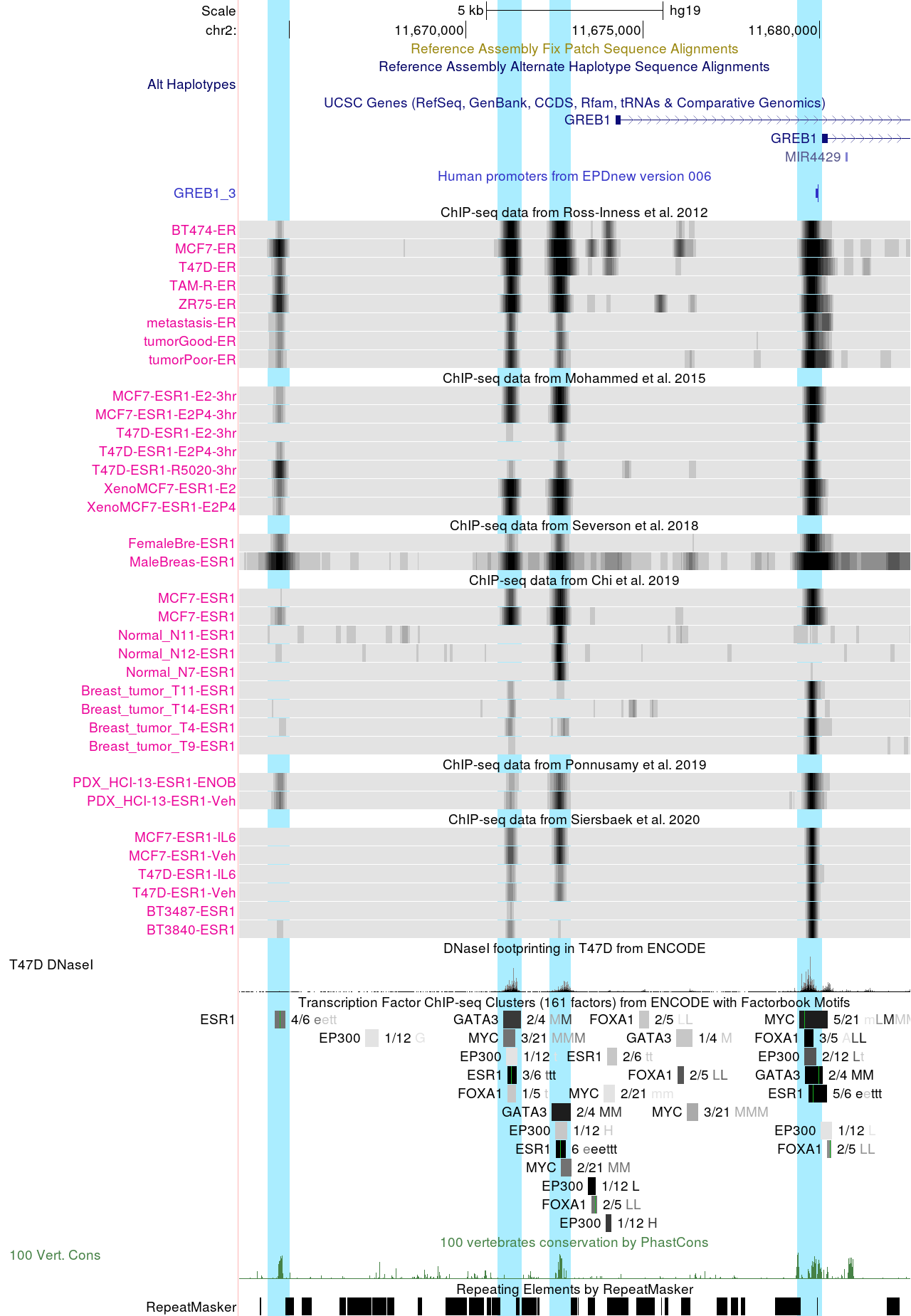
This first set of sessions are customized views focusing on ESR1 binding sites at classical estrogen-regulated genes such as GREB1, CCND1, PGR, and MYC. ESR1 binding is also shown at the CDH1 enhancer region. CDH1 downregulation is positively associated with epithelial-to-mesenchymal transition (EMT) during initial tumor invasion.
We also display data showing how up-regulation of AIM2, a tumor suppressor gene, is possibly mediated by cytokine-induced pSTAT3. The session link to the ZEB1 gene, a key player in EMT progression, shows how IL6-induced pSTAT3 binding could regulate ZEB1 transcription in breast cancer cells (MCF7, and T47D).
Finally, we also provide two browser sessions displaying PGR and AR binding in enhancer regions of the KLK3 and RANKL genes.
In the Session Gallery (on the left-side menu under the Documentation section), for each screenshot, we provide an accompanying text that explains the biological background and walks users through the pictures.
Breast Cancer Epigenomics Track Hub Viewer for H. sapiens (Feb 2009 GRCh37/hg19), version 1.0
Here below is a table summarizing the track content of the hub (latest version). Note that when strand is n/a, it means that signal applies to both strands.